The Structure Lab
Visualizing the Intricacies of Protein Architecture
At KACTUS, we're committed to integrating accurate protein structure into every product we offer. Our Structure Lab embodies this commitment, where we create detailed 3D protein models that showcase the intricate architecture of these essential molecules. These models are a testament to our dedication to providing researchers with physiologically relevant and bioactive proteins for drug discovery.
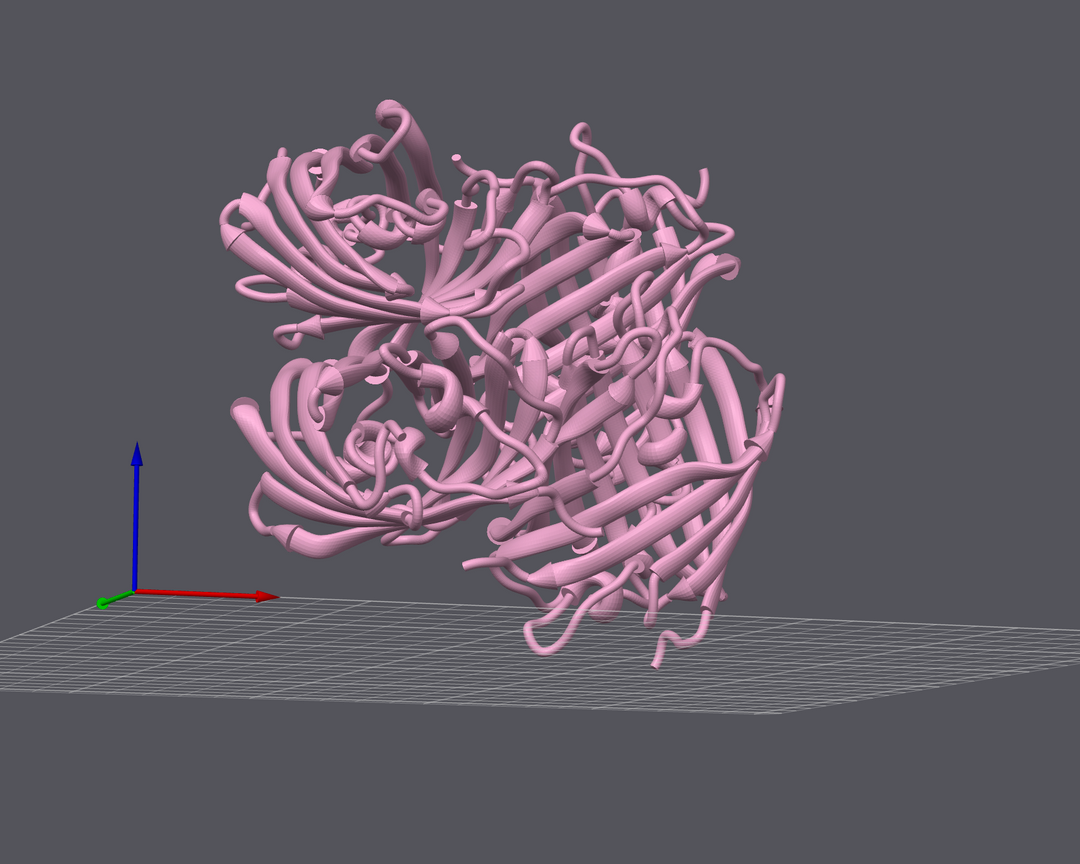
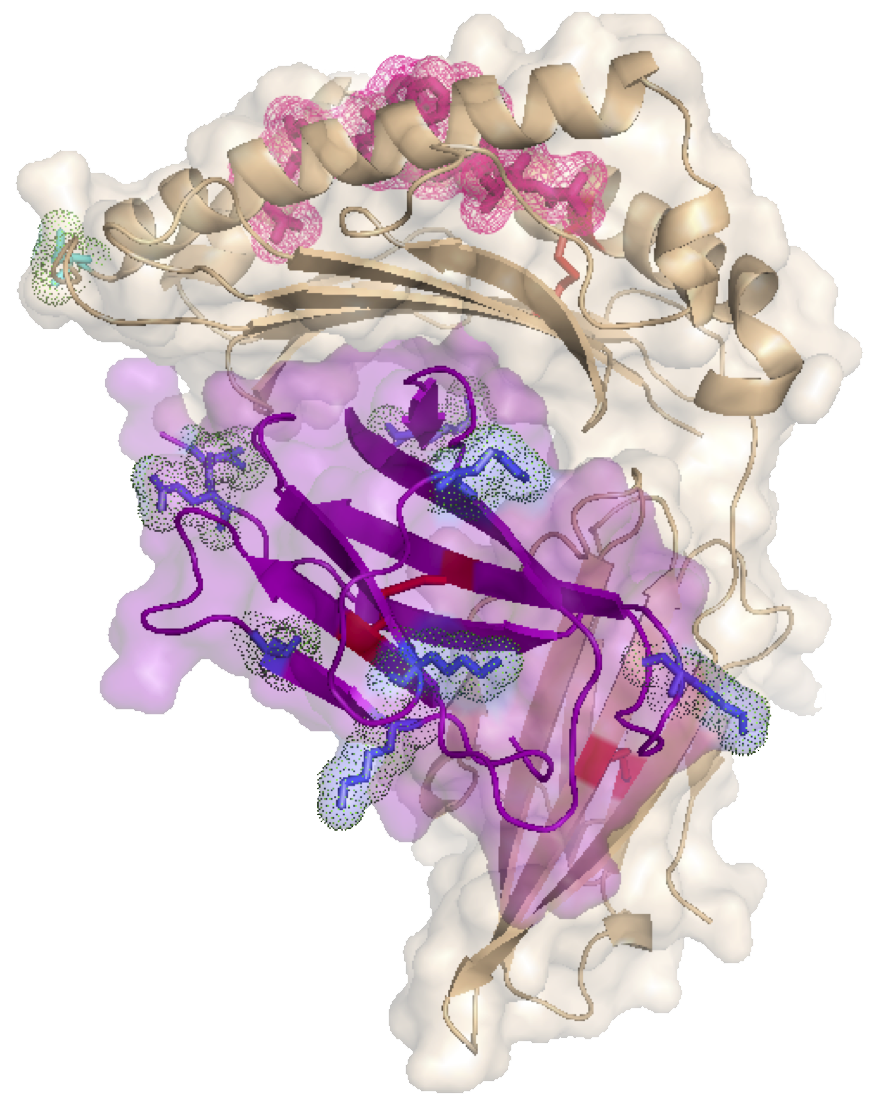
Why Protein Structure Matters
Structure Meets Function
Proteins are the workhorses of life, carrying out a vast array of functions in every living organism. A protein's structure dictates how it interacts with other molecules, influencing its ability to bind to targets, catalyze reactions, and participate in signaling pathways. Even minor alterations in structure can significantly impact a protein's activity and behavior.
Utilizing accurate recombinant proteins is paramount to effective biopharmaceutical development.
Our Commitment to Structural Accuracy
We recognize the critical importance of accurate protein structure in biopharmaceutical development.
Our team of experts employs rigorous study of the 3D protein structure to ensure the bioactivity, reliability, and physiological-relevance of our proteins.
By understanding the intricate details of protein structure, we strive to accelerate effective and efficient drug discovery.
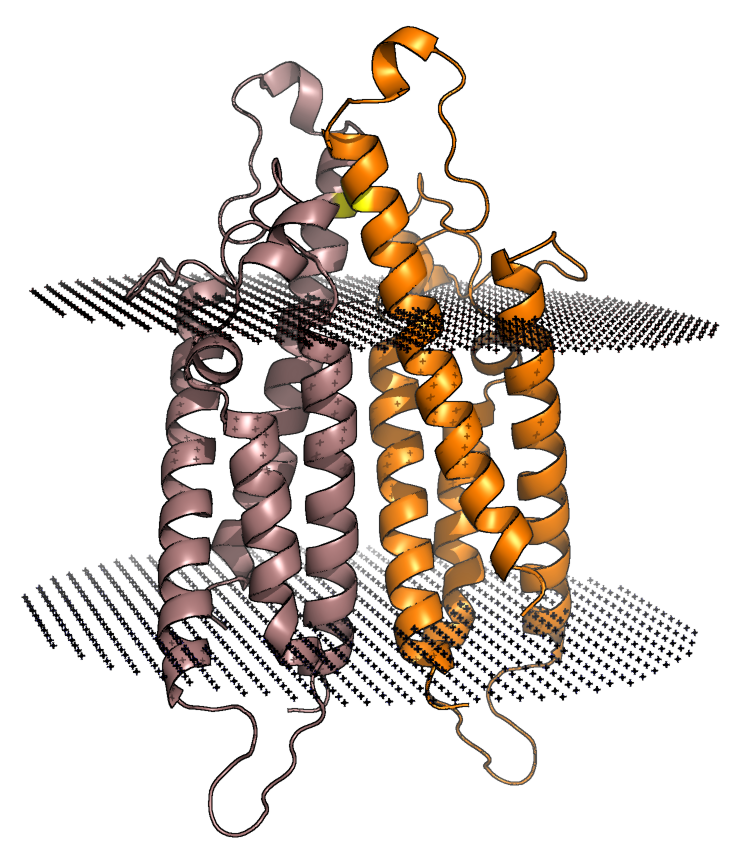
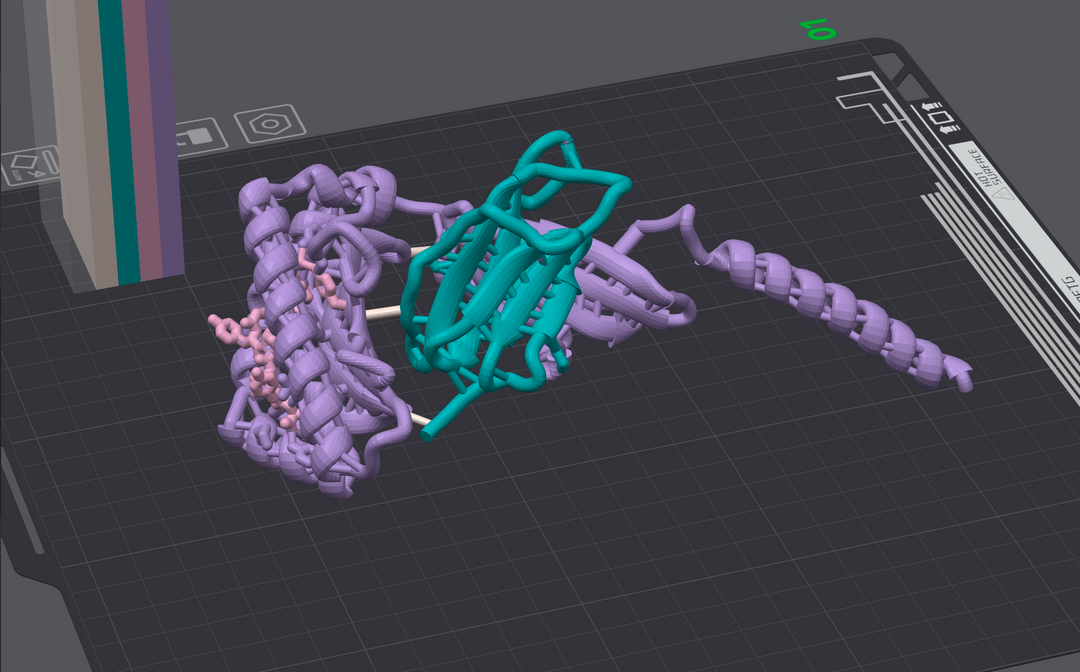
3D Printing Protein Architecture
To truly appreciate the complexity of protein structure, we've brought the invisible to life. Using cutting-edge 3D printing technology, we transform digital structural data into tangible models that you can hold in your hand.
These models provide a unique perspective on protein architecture, allowing researchers to visualize intricate folds, binding sites, and surface features in a way that traditional representations cannot. By bridging the gap between the digital and physical worlds, we aim to deepen understanding and inspire new breakthroughs in drug discovery.
Visualizing KACTUS Recombinant Proteins
Each protein model in our collection corresponds to one of the recombinant proteins in our catalog, offering you a tangible glimpse into their structure. Browse some examples below.
MHC Class I
An MHC Class I molecule crucial for the immune system to recognize and destroy infected or cancerous cells.
TCR-CD3 Complex
This complex represents the interaction between a T cell receptor and a CD3 protein, a critical component of the immune response.
3D Protein Structure FAQs
The Structure Lab specializes in creating 3D models of protein structures. It supports research and drug discovery by providing accurate visualizations of protein folding, binding sites, and molecular architecture relevant to biological function.
A protein’s structure determines how it interacts with other molecules. Structural insights help researchers predict binding affinities, assess conformational stability, and design targeted therapeutics with improved specificity and efficacy.
Protein structures are analyzed using a combination of computational modeling, structural databases, and experimental data from sources like X-ray crystallography or cryo-electron microscopy.
Digital models are generated using structural informatics and bioinformatics tools. These are then converted into tangible 3D-printed representations to visualize folds, domains, and active sites.
3D-printed models offer a physical, tactile perspective of protein architecture, allowing researchers to better understand spatial relationships and surface features that influence biological activity.
Examples include MHC Class I, CCR8, and the TCR-CD3 complex. Each model corresponds to a recombinant protein offered in the catalog and illustrates relevant functional domains.
Structural accuracy ensures that recombinant proteins mimic their native conformation, which is essential for proper folding, target binding, and functional assays in biopharmaceutical development.
The lab aims to bridge digital structure prediction with hands-on scientific insight. Its mission is to deepen understanding of protein behavior and accelerate translational research in the life sciences.







