Background
In recent years, the field of cell and gene therapy has made rapid progress. It is reported that there are over 300 clinical projects based on AAV vectors worldwide, generally produced through triple plasmid co-transfection. Additionally, there are nearly 1,000 CAR-T clinical trials underway, most of which transduce CAR constructs using lentiviral vectors produced through transient transfection using four plasmids. Consequently, the demand for plasmids in the market is also continuously increasing.
Traditional plasmid production involves the fermentation of E. coli, which poses several challenges in commercial production. For instance, plasmids contain extraneous sequences, such as antibiotic resistance genes and origins of replication, which can affect plasmid yield. The entire process, which includes fermentation, extraction, purification, and quality control, is time-consuming and costly. Moreover, there is a risk of genetic recombination between E. coli and plasmid DNA, leading to safety incidents.
In vitro enzymatic synthesis of DNA vectors can effectively replace traditional plasmid DNA production, perfectly avoiding the many uncontrollable risks of biological fermentation. The production process is simpler and more controllable, allowing for commercial production in a shorter time frame and having broad applications in the field of cell and gene therapy.
KACTUS has developed a series of off-the-shelf, high-activity enzyme for in vitro synthesis of DNA vectors, based on our functional recombinant protein and enzyme technology platform, SAMS™.
Product Features & Performance
TelN Protelomerase
TelN protelomerase cleaves dsDNA at its recognition site and leaves covalently closed ends. It can be used for DNA vaccine development as well as for non-viral therapy vectors. TelN Protelomerase is isolated from phage N15.
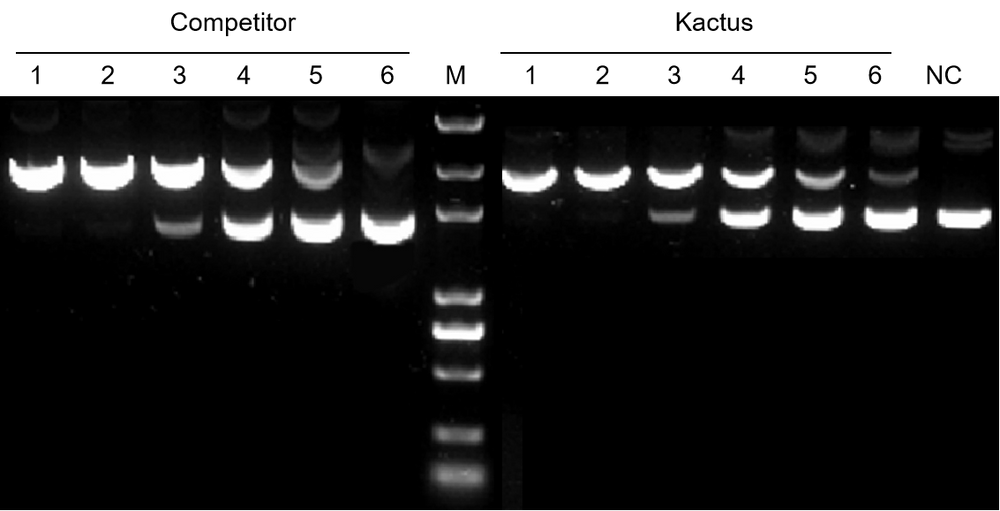
Figure 1. TelN Protelomerase efficiently converts plasmid DNA into linear DNA with hairpin ends. In this figure, TelN Protelomerase cut substrate DNA containing 513 fmol cleaveage site, followed by agarose gel analysis.
Phi29 DNA Polymerase
Phi29 DNA polymerase is a highly processive enzyme that can efficiently amplify DNA sequences in vitro. It is often used as an alternative to the more commonly used polymerase chain reaction (PCR) method for DNA amplification. Phi29 DNA polymerase amplification has several advantages over PCR, including higher processivity, reduced reaction time, and higher fidelity. It is particularly useful for amplifying DNA from samples with limited starting material, such as ancient DNA or single cells. It is a highly robust enzyme and is suitable for rolling circle amplification (RCA), multiple-displacement amplification (MDA), and whole genome amplification.
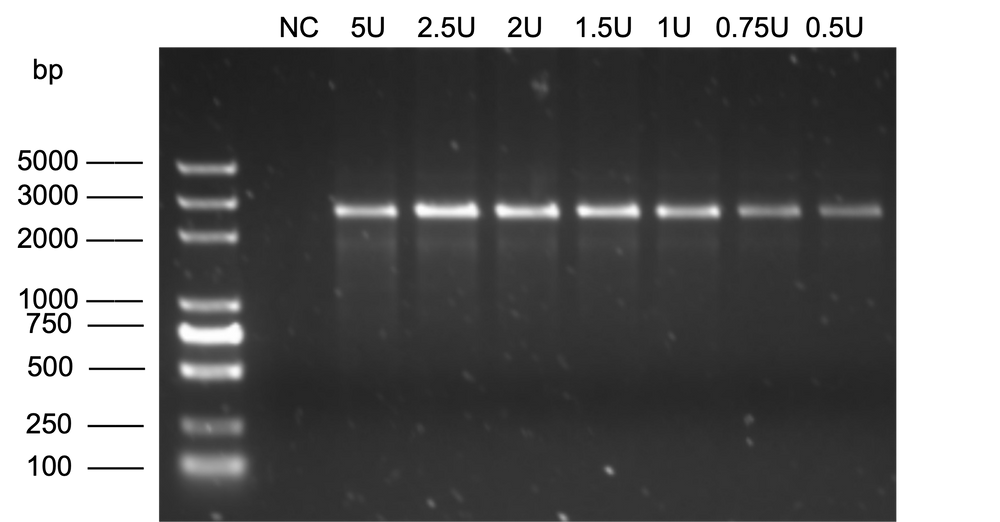
Figure 2. Rolling cycle amplification of pUC57 plasmid with phi29 DNA Polymerase. Product was digested with BspQI and then analyzed with Agarose gel.
Exonuclease III
Exonuclease III is a highly processive enzyme that hydrolyzes the phosphodiester bonds in DNA in the 3' to 5' direction, resulting in stepwise removal of nucleotides. Exonuclease III catalyzes the removal of nucleotides from linear or nicked double stranded DNA. Degradation of Exonuclease III could be initiated from a 3’ blunt end, 3’ recessed end, 3’ overhangs with less than 4 bases and nicked DNA.
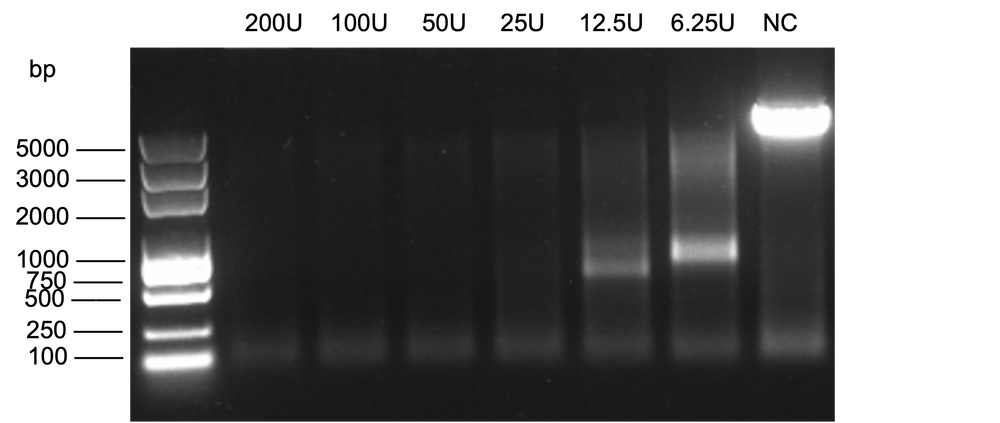
Figure 3. Digestion of 1μg linearized plasmid using Exonuclease III, followed by Agarose gel analysis.
Restriction Endonucleases
There are two types of DNA vectors formed by TelN Protelomerase cutting and ligation, one containing the backbone sequence and the other containing the target fragment. There are corresponding restriction enzyme sites on the backbone sequence. KACTUS provides highly active restriction endonucleases for digestion of the backbone sequence.
| Enzyme | Cut Site |
|---|---|
| BsaI | 5'...GGTCTC(N)1 ↓ ...3' 3'...CCAGAG(N)5 ↑ ...5' |
| BspQI | 5'...GCTCTTC(N)1 ↓ ...3' 3'...CGAGAAG(N)4 ↑ ...5' |
| KpnI | 5'...GGTAC ↓ C...3' 3'...C ↑ CATGG...5' |
| NruI | 5'...TCG ↓ CGA...3' 3'...AGC ↑ GCT...5' |
| SalI | 5'...G ↓ TCGAC...3' 3'...CAGCT ↑ G...5' |
| SapI | 5'...GCTCTTC(N)1 ↓ ...3' 3'...CGAGAAG(N)4 ↑ ...5' |
| SapI V2.0 | 5'...GCTCTTC(N)1 ↓ ...3' 3'...CGAGAAG(N)4 ↑ ...5' |
| XbaI | 5'...T ↓ CTAGA...3' 3'...AGATC ↑ T...5' |
Applications
- Preparation of lentiviral vectors
- Preparation of AAV vectors
- Preparation of DNA templates in the early stage of DNA vaccine production
- Preparation of DNA template in the early stage of mRNA vaccine production

About KACTUS

Quality Control

SAMS™ Technology Platform
DNA Amplification & Modification Enzymes FAQs
Enzymatic DNA synthesis eliminates the need for bacterial fermentation, avoiding antibiotic resistance markers, origins of replication, and risk of recombination. It also shortens timelines by removing cell lysis and purification steps.
TelN Protelomerase cleaves double-stranded DNA at a specific recognition site and generates covalently closed hairpin ends. This creates linear DNA vectors suitable for vaccine and non-viral therapy applications.
Phi29 DNA Polymerase offers high processivity and strand displacement activity, enabling whole-genome amplification and isothermal methods such as rolling circle amplification. It does not require thermal cycling.
Phi29 is ideal for multiple displacement amplification (MDA), whole genome amplification, single-cell genomics, and rolling circle amplification of plasmids or circular DNA templates.
Exonuclease III removes nucleotides in a 3’ to 5’ direction from double-stranded DNA. It is useful for creating nested deletions or processing linear or nicked DNA with recessed or blunt 3’ ends.
KACTUS provides restriction enzymes including BsaI, BspQI, KpnI, NruI, SalI, SapI, and XbaI, which are commonly used for template linearization, backbone digestion, or fragment release in DNA assembly and cloning workflows.
Yes. The enzymes are suitable for preparing DNA templates used in plasmid-based vector production for lentivirus and AAV systems, especially during transient transfection steps.
TelN Protelomerase-generated linear vectors are used in non-viral gene delivery systems, DNA vaccines, and cases where circular plasmids introduce undesirable genetic elements or complexity.
Each enzyme is evaluated for activity using substrate-specific assays, such as digestion patterns on agarose gels or amplification yields, ensuring consistent lot-to-lot performance.