Base Editing Technology Leads a New Chapter in Drug Development
By Yujiao Zhang
Since its debut in 2016, base editing technology has opened up new perspectives and methods for drug development with its outstanding precision and reliability. These advances rely heavily on high-quality gene editing enzymes that ensure accuracy, safety, and scalability for clinical applications. This year, the clinical data of two base editing drug candidates will be unveiled, which is crucial for validating the clinical application of base editing technology and has attracted widespread attention in the industry.
Verve-101
Nature Medicine lists 11 key clinical trials in 2024 that will significantly impact the medical field [1]. Among all of them, VERVE-101 is an in vivo base editing drug that targets the inactivation of the PCSK9 gene in the liver to persistently lower the level of low-density lipoprotein cholesterol (LDL-C) in the blood, treating familial hypercholesterolemia. Verve-101 received FDA clearance of IND application in October 2023.
Beam-101
The Beam-101, developed by Beam Therapeutics, was included by the industry media BioPharma Dive in the 10 clinical trials that could significantly impact the industry in the first half of this year [2]. Beam-101 is an ex vivo single base editing therapy that alleviates sickle cell disease by editing patients' hematopoietic stem cells to increase fetal hemoglobin expression. This therapy integrates gene editing and cell therapy and received FDA clearance of IND application in November 2021.
![Examples of gene editing drugs in clinical use [3].](https://cdn.shopify.com/s/files/1/0627/6025/5710/files/Fig_1_2f23aae1-a2fb-4648-9557-ba0dfc3f0554_480x480.png?v=1707935831)
Figure 1. Examples of gene editing drugs in clinical use [3].
Advantages of Base Editing
- Accurate gene modification through precise base substitution without inducing DNA double-strand breaks. For example, the cytosine base editor (CBE) can convert the base pair C•G into T•A, while the adenine base editor (ABE) can alter the base pair A•T into G•C;
- No chromosome translocations, deletions, or p53 activation as a result of DNA double-strand breaks, much safer for treatments.
- Capable of editing both dividing and non-dividing cells;
- High gene editing efficiency observed in various cells types: Over 80% editing in hematopoietic stem cells (HSCs), over 90% editing in primary T cells, and over 80% editing in induced pluripotent stem cells (iPSCs) [4].
Applications of Base Editing
Base editing technology has a wide range of applications. It can either precisely mutate a single base to correct genetic defects caused by single base mutations or delete genes by introducing stop codons, mutating splicing sites, etc (see below).

Figure 2. The application of base editing technology (using CBE as an example).
-
Reversing disease-causative point mutations
Single nucleotide polymorphisms (SNPs) are the most common type of mutation associated with human diseases, with 70% of these mutations potentially correctable through base editing. This sheds light on a new pathway to rectify genetic defects [5].
β-thalassemia is a blood genetic disorder caused by mutations in the HBB gene. The HBB-28 (A>G) mutation is one of the top 3 common mutations observed in the patient population in China and Southeast Asia. It has been reported that using base editors (BEs) to correct the HBB-28 (A>G) mutation in patient-derived primary cells were able to achieve an editing efficiency of over 20% [6].
Base editing can also be used to reverse pathogenic mutations in patient-derived induced pluripotent stem cells (iPSCs), achieving an editing efficiency of over 80% These iPSCs can be applied in disease mechanism studies, drug screening, and cell therapy development [7].
-
Introducing stop codons for Gene Knockout
Cytosine base editors (CBEs) can convert C•G base pairs into T•A, introducing a premature stop codon to disrupt gene expression. Substituting cytosines in coding sequences to thymine can generate stop codons like TAA, TAG, or TGA. This method has been reported to have similar efficiency to Cas9-mediated gene knockouts [8].
-
Disrupting RNA splicing sites for Gene Knockout
Gene knockout can also be achieved by disrupting mRNA splicing sites such as splice donor (SD) sites and, potentially, more advantageous over introducing stop codons directly, as altering the splicing site can avoid issues with stop codon reading under stress conditions. Editing RNA splicing sites changes gene processing at the mRNA level and therefore, blocks protein translation.
An example of applying base editing technology in gene knockout includes the knockout of TRAC, B2M, & PD1 genes to create allogenic CAR-T cells—an approach increasingly relevant in antibody drug discovery. Delivering base editors in the form of ribonucleoproteins (RNPs) via electroporation for triple gene knockouts achieved over 90% gene knockout efficiency in primary T cells [9].
The VERVE-101 leverages an adenine base editor to alter the PCSK9 mRNA splicing site, leading to aberrant splicing and nonsense-mediated decay, thereby reducing PCSK9 protein levels. These examples demonstrate the versatility and potential of base editing technology in creating precise genetic modifications for therapeutic purposes, offering hope for treating a wide range of genetic disorders.
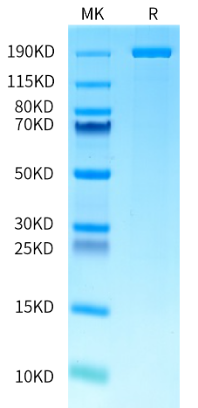
The AccuBase™ “near-zero off-target” base editing tools
After a decade of competition, gene editing technology has developed into the 2.0 phase represented by base editors [10]. The CBE base editing technology AccubaseTM, independently developed and owned by Base Therapeutics, has been validated for its high-efficiency and precision gene editing capabilities across various cells and applications. In 2023, Base Therapeutics joined forces with KACTUS in a strategic partnership for the global production and sales of the GMP-grade base editor AccuBaseTM. Today, the AccuBaseTM products are globally available in stock, supporting CGT drug development.
Key achievements with AccuBase™ include:
High-efficiency gene knockout in activated primary T cells using AccuBase™ RNP, achieving over 80% knockout efficiency for PD1 and B2M, and 96% for TRAC.
Nearly 70% triple-gene knockout efficiency in NK cells using AccuBase™ RNP.

"Near-zero off-target" effects. Using GOTI (Genome-wide Off-target analysis by Two-cell embryo Injection), a method that evaluates the whole-genome off-target levels by comparing the edited cells to a control group. The comparability study shows that the single nucleotide variants (SNVs) occurring in the AccuBase™ group were found to be close to the level of the GFP control group, approaching zero, while the control base editor group has significant high level of SNV occurrence.

Product Information for AccuBase™
KACTUS provides both Research-Grade and GMP-grade AccuBase™ products on the market, which are currently in stock. Feel free to reach out and request a testing sample today!
|
Product Name |
Catalog |
Size |
|
KD-0001 |
200μg/500μg/1mg |
|
|
GMP-KD-0001 |
1mg |
NOTE: BS-EP1 is product name, AccuBase™ is the commercial name.
References
[1] 11 clinical trials that will shape medicine in 2024. Nat Med 29, 2964–2968 (2023).
[2] BioPharma Dive: 10 clinical trials to watch in the first half of 2024.
[3] In the business of base editors: Evolution from bench to bedside. PLoS Biol. 2023 Apr 12;21(4):e3002071.
[4] In vivo somatic cell base editing and prime editing. Mol Ther. 2021 Nov 3;29(11):3107-3124.
[5] Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018 Dec;19(12):770-788.
[6] Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell. 2017 Nov;8(11):811-822.
[7] Efficient Generation and Correction of Mutations in Human iPS Cells Utilizing mRNAs of CRISPR Base Editors and Prime Editors. Genes (Basel). 2020 May 6;11(5):511.
[8] CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017 Jul;14(7):710-712.
[9] Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat Commun. 2019 Nov 19;10(1):5222.
[10] CRISPR 2.0: a new wave of gene editors’ heads for clinical trials. Nature. 2023 Dec;624(7991):234-235.